Ubiquitination
Introduction
Ubiquitination is a post-translational modification process that involves the attachment of ubiquitin, a small regulatory protein, to a substrate protein. This process plays a crucial role in various cellular functions, including protein degradation, cell cycle regulation, DNA repair, and signal transduction. Ubiquitination is a highly regulated and complex process that ensures cellular homeostasis and proper functioning.
Ubiquitin and Its Structure
Ubiquitin is a small protein consisting of 76 amino acids and is highly conserved across eukaryotic species. It has a compact, globular structure with a β-grasp fold, which is stabilized by hydrophobic interactions and hydrogen bonds. The C-terminal glycine residue of ubiquitin is crucial for its conjugation to substrate proteins.
The Ubiquitination Process
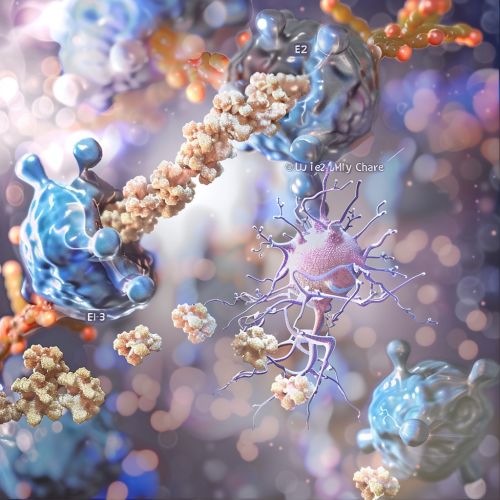
The ubiquitination process involves three main enzymatic steps: activation, conjugation, and ligation.
E1: Ubiquitin-Activating Enzyme
The ubiquitination process begins with the activation of ubiquitin by an E1 ubiquitin-activating enzyme. This enzyme catalyzes the adenylation of the C-terminal glycine of ubiquitin, forming a ubiquitin-adenylate intermediate. Subsequently, a thioester bond is formed between the C-terminal glycine of ubiquitin and a cysteine residue on the E1 enzyme.
E2: Ubiquitin-Conjugating Enzyme
The activated ubiquitin is then transferred to an E2 ubiquitin-conjugating enzyme via a trans-thioesterification reaction. The E2 enzyme plays a pivotal role in determining the specificity of substrate recognition and the type of ubiquitin chain formed.
E3: Ubiquitin Ligase
The final step involves the transfer of ubiquitin from the E2 enzyme to the substrate protein, a process facilitated by an E3 ubiquitin ligase. E3 ligases confer substrate specificity and catalyze the formation of an isopeptide bond between the C-terminal glycine of ubiquitin and the ε-amino group of a lysine residue on the substrate protein.
Types of Ubiquitination
Ubiquitination can occur in various forms, including monoubiquitination, multi-monoubiquitination, and polyubiquitination.
Monoubiquitination
Monoubiquitination involves the attachment of a single ubiquitin molecule to a substrate protein. This modification is often involved in processes such as endocytosis, DNA repair, and histone regulation.
Multi-monoubiquitination
Multi-monoubiquitination refers to the attachment of single ubiquitin molecules to multiple lysine residues on the same substrate protein. This type of ubiquitination is implicated in receptor internalization and trafficking.
Polyubiquitination
Polyubiquitination involves the formation of ubiquitin chains on a substrate protein. These chains can be linked through different lysine residues on ubiquitin, leading to various functional outcomes. The most well-studied polyubiquitin chains are linked through lysine 48 (K48) and lysine 63 (K63).
Functional Implications of Ubiquitination
Ubiquitination serves as a versatile regulatory mechanism in numerous cellular processes.
Protein Degradation
One of the primary functions of ubiquitination is to target proteins for degradation via the proteasome. K48-linked polyubiquitin chains are recognized by the 26S proteasome, leading to the degradation of the substrate protein.
DNA Repair
Ubiquitination plays a critical role in the DNA damage response. For instance, the E3 ligase BRCA1 mediates the ubiquitination of histones and other proteins involved in DNA repair pathways, facilitating the recruitment of repair factors to sites of damage.
Cell Cycle Regulation
Ubiquitination regulates the cell cycle by controlling the stability of key cell cycle regulators. The APC/C is an E3 ligase that targets cell cycle proteins such as cyclins and securin for degradation, ensuring proper cell cycle progression.
Signal Transduction
Ubiquitination modulates various signaling pathways, including the NF-κB pathway. The E3 ligase TRAF6 mediates K63-linked polyubiquitination of signaling proteins, promoting the activation of downstream effectors.
Deubiquitination
Deubiquitination is the process of removing ubiquitin from substrate proteins, a reaction catalyzed by deubiquitinating enzymes (DUBs). DUBs play a crucial role in maintaining the balance of ubiquitination and ensuring the reversibility of ubiquitin-mediated processes.
Clinical Relevance
Dysregulation of ubiquitination is implicated in various diseases, including cancer, neurodegenerative disorders, and immune system dysfunctions. For example, mutations in the E3 ligase Parkin are associated with Parkinson's disease, highlighting the importance of ubiquitination in neuronal health.
Research and Therapeutic Approaches
Research on ubiquitination has led to the development of therapeutic strategies targeting the ubiquitin-proteasome system. Proteasome inhibitors, such as bortezomib, are used in the treatment of multiple myeloma, demonstrating the potential of targeting ubiquitination pathways in disease management.